State-of-the-Science Stories: Functional Genomics
What is a Functional Genomics?
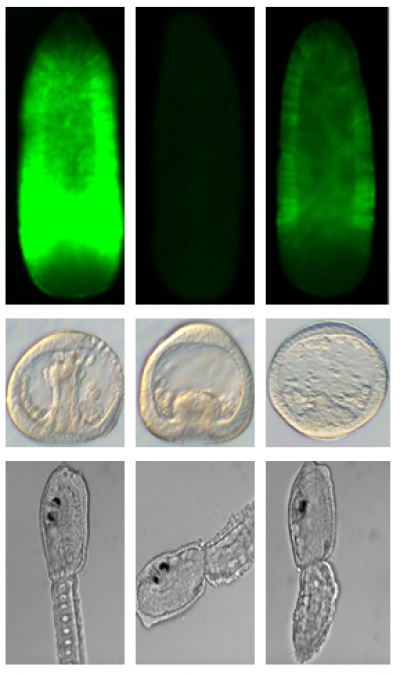
Functional genomics is a field of molecular biology that attempts to describe gene (and protein) functions and interactions.
These days it is quite easy to obtain whole genome or transcriptome data and much research focusses on this. The study of the specific functions of single genes is also a vital part of understanding genetics, but it is difficult to do compared genome-wide studies. This is therefore the focus of JRA3's work: single-gene function studies using up-to-date approaches such as CRISPR-Cas9. ASSEMBLE Plus will adapt and implement protocols to generate genetically modified metazoans, macroalgae and microorganisms, all of which represent strategically important resources for academia and some of them also for industry (e.g. bio-actives for food, feed, pharma and cosmetics).
What is ASSEMBLE Plus doing that is different?
CRISPR is a simple yet powerful tool that is used for editing genomes. It allows researchers to easily alter DNA sequences and modify gene function. Its many potential applications include correcting genetic defects, studying gene functions, treating and preventing the spread of diseases, and improving crops through transgenesis.
For the purpose of our ASSEMBLE Plus work, CRISPR can be used to knock out a gene, to then study the effect of its loss and therefore its function. The protocols for doing this have been fully established for the sea urchin (both Paracentrotus lividus and Strongylocentrotus purpuratus) and three other marine metazoans (the cnidarian Clytia hemisphaerica and the ascidians Ciona intestinalis, and Phallusia mammillata). However, in amphioxus (the cephalochordate Branchiostoma lanceolatum), even if the first results are promising now that the JRA3 team has shown that CRISPR-Cas9 can mutate genes in these cephalochordate embryos, not all cells mutate in the first generation and so more work is necessary to obtain complete mutant lineages. Other approaches such as insertional transgenesis worked in all the four animal models, sea urchins, ascidians, Clytia and amphioxus, and also with different ratios of insertion, from complete mutants to mosaic animals.
All the required material is ready to explore the brown algae Ectocarpus. Work is underway on the diatom Phaeodactylum tricornutum, with some gene knockout strains identified.
What next?
- Work will continue on macroalgaes including Ectocarpus and kelps, especially Saccharina latissima).
- Work will continue on microorganisms including (the diatoms Seminavis, Cylindrotheca and Pseudo-nitzschia multistriata, the picoeukaryotes Ostreococcus sp, Bathycoccus and Micromonas, the cyanobacteria Synechococcus and the bacteria Kordia algicida).
Potential Impact
Once ASSEMBLE Plus gains a better understanding of the functional genomics of the species covered, it will be possible to study many different gene functions, and the bioactive properties of some of their gene products.
Protocols for genetic transformation and generation of knockouts will be produced. This project will continue after ASSEMBLE Plus. So far we have achieved good results for both macroalgae and diatoms. For now we are concentrating on targeting academia but in the future there is the possibility to use these protocols on other organisms. JRA3 has published many papers covering the protocols (see the ASSEMBLE Plus publications collection) and two deliverables - D9.1: Protocols for genetic transformation of model organisms and D9.2: Protocols for the deployment of CRISPRCas9 system.


